rdhs client object and internal package design
OJ Watson, Jeff Eaton
2018-09-24
Source:vignettes/client.Rmd
client.RmdOverview
The following is a similar vignette - “How to use rdhs?”. However,
this one achieves the same results using the rdhs client
object directly to download datasets etc, rather than using the easier
user interfacing functions. This vignette is included to help
demonstrate how the pacakge internals are working and adds more
information about how the package was designed, which may be useful for
anyone adding extra functionality to rdhs in the
future.
If are not bothered by this then please read the main introductory vignette.
rdhs is a package for management and analysis of Demographic and Health Survey
(DHS) data. This includes functionality to:
- Access standard indicator data (i.e. DHS STATcompiler) in R via the DHS API.
- Identify surveys and datasets relevant to a particular analysis.
- Download survey datasets from the DHS website.
- Load datasets and associated metadata into R.
- Extract variables and combining datasets for pooled multi-survey analyses.
This process is described below and should cover most functionality that will be needed for working with these datasets.
1. Access standard indicator data via the API
The DHS API has many endpoints that can be accessed using
anyone of dhs_<endpoint>() functions. All exported
functions within rdhs that start dhs_ interact
with a different with a different endpoint of the DHS API. Their website gives
great information about the different search terms and filters that can
be used, and we have tried to include all of this within the
documentation of each function.
One of those endpoints, dhs_data(), interacts with the
the published set of standard health indicator data calculated by the
DHS. This endpoint allows use to retrieve a set of health indicators
that have been sample weighted to give country, subnational estimates
that can be further refined by education and wealth brackets. To do this
we use the dhs_data() endpoint, which we can then either
search for specific indicators, or by querying for indicators that have
been tagged within specific areas.
## what are the indicators
indicators <- dhs_indicators()
indicators[1,]## Definition
## 1 Age-specific fertility rate for the three years preceding the survey for age group 10-14 expressed per 1,000 women
## NumberScale IndicatorType MeasurementType IsQuickStat ShortName
## 1 0 I Rate 0 ASFR 10-14
## IndicatorId Level1 IndicatorTotalId Level2 Level3
## 1 FE_FRTR_W_A10 Fertility Fertility rates Women
## SDRID IndicatorOldId TagIds DenominatorWeightedId
## 1 FEFRTRWA10
## Label IndicatorOrder
## 1 Age specific fertility rate: 10-14 11763005
## Denominator
## 1 Per thousand women years exposed in the period 1-36 months prior to interview
## QuickStatOrder IndicatorSpecial1Id DenominatorUnweightedId
## 1
## IndicatorSpecial2Id
## 1Each call to a DHS endpoint returns a data.frame by
default with all the results available by default.
The DHS has a unique IndicatorId for each of the statistics it calculates. The definition and specific string for each indicator is included within the IndicatorId and Definition variable:
# grab the first 5 alphabetically
indicators[order(indicators$IndicatorId),][1:5,c("IndicatorId", "Definition")]## IndicatorId
## 2031 AH_CIGA_M_UNW
## 2026 AH_CIGA_W_10P
## 2023 AH_CIGA_W_12C
## 2024 AH_CIGA_W_35C
## 2025 AH_CIGA_W_69C
## Definition
## 2031 Number of men who smoke cigarettes (unweighted)
## 2026 Percentage of women who smoked 10+ cigarettes in preceding 24 hours
## 2023 Percentage of women who smoked 1-2 cigarettes in preceding 24 hours
## 2024 Percentage of women who smoked 3-5 cigarettes in preceding 24 hours
## 2025 Percentage of women who smoked 6-9 cigarettes in preceding 24 hoursSince there are quite a lot of indicators, it might be easier to
first query by tags. The DHS tags their indicators by what areas of
demography and health they relate to, e.g. anaemia, literacy, malaria
parasitaemia are all specific tags. First let’s look at what the tags
are, by interacting with the dhs_tags() endpoint, before
grabbing data that related to malaria parasitaemia in the DRC and
Tanzania since 2010:
# What are the tags
tags <- dhs_tags()
# Let's say we want to view the tags that relate to malaria
tags[grepl("Malaria", tags$TagName), ]## TagType TagName TagID TagOrder
## 31 0 Malaria Parasitemia 36 540
## 43 2 Select Malaria Indicators 79 1000
# and now let's then grab this data by specifying the countryIds and the survey year starts
data <- dhs_data(tagIds = 36,countryIds = c("CD","TZ"),breakdown="subnational",surveyYearStart = 2010)
data[1,]## DataId Indicator SurveyId IsPreferred Value
## 1 941297 Malaria prevalence according to RDT CD2013DHS 1 17.1
## SDRID Precision RegionId SurveyYearLabel SurveyType
## 1 MLPMALCRDT 1 CDDHS2013503010 2013-14 DHS
## SurveyYear IndicatorOrder DHS_CountryCode CILow
## 1 2013 125136010 CD
## CountryName IndicatorType CharacteristicId
## 1 Congo Democratic Republic I 503010
## CharacteristicCategory IndicatorId CharacteristicOrder
## 1 Region ML_PMAL_C_RDT 1503010
## CharacteristicLabel ByVariableLabel DenominatorUnweighted
## 1 Kinshasa 406
## DenominatorWeighted CIHigh IsTotal ByVariableId
## 1 532 0 0Depending on your analysis this maybe more than enough detail. It is
also worth mentioning that this data can also be accessed via DHS STATcompiler if you prefer
a click and collect version. However, hopefully one can see that
selecting a lot of different indicators for multiple countries and
breakdowns should be a lot easier using the rdhs API
interaction. For example we can very quickly find out the trends in
antimalarial use in Africa, and see if perhaps antimalarial prescription
has decreased after RDTs were introduced (assumed 2010).
# Make an api request
resp <- dhs_data(indicatorIds = "ML_FEVT_C_AML", surveyYearStart = 2010,breakdown = "subnational")
# filter it to 12 countries for space
countries <- c("Angola","Ghana","Kenya","Liberia",
"Madagascar","Mali","Malawi","Nigeria",
"Rwanda","Sierra Leone","Senegal","Tanzania")
# and plot the results
library(ggplot2)
ggplot(resp[resp$CountryName %in% countries,],
aes(x=SurveyYear,y=Value,colour=CountryName)) +
geom_point() +
geom_smooth(method = "glm") +
theme(axis.text.x = element_text(angle = 90, vjust = .5)) +
ylab(resp$Indicator[1]) +
facet_wrap(~CountryName,ncol = 6) 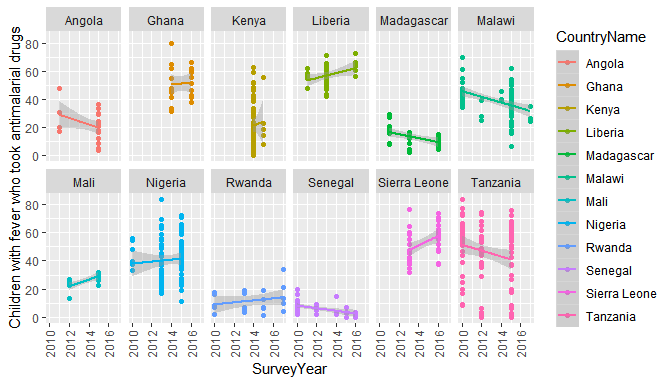
If we incorrectly entered a filter query (very possible),
rdhs will let us know our request was invalid:
# Make an api request
resp <- dhs_data(indicatorIds="ML_FEVT_C_AMasfafasfL",
surveyYearStart=202231231306,
breakdown="subParTyping")## Error in handle_api_response(resp, TRUE):
## -> DHS API Request Failed [500]
## -> Error Type: dhs_internal_server_error2. Identify surveys relevant for further analysis
You may, however, wish to do more nuanced analysis than the API allows. The following 4 section detail a very basic example of how to quickly identify, download and extract datasets you are interested in.
Let’s say we want to get all the survey data from the Democratic Republic of Congo and Tanzania in the last 5 years (since 2013), which covers the use of rapid diagnostic tests (RDTs) for malaria. To begin we’ll interact with the DHS API to identify our datasets.
To start our extraction we’ll query the
surveyCharacteristics endpoint using
dhs_surveyCharacteristics():
## make a call with no arguments
sc <- dhs_survey_characteristics()
sc[grepl("Malaria", sc$SurveyCharacteristicName), ]## SurveyCharacteristicID SurveyCharacteristicName
## 57 96 Malaria - DBS
## 58 90 Malaria - Microscopy
## 59 89 Malaria - RDT
## 60 57 Malaria module
## 61 8 Malaria/bednet questionsThere are 87 different survey characteristics, with one specific
survey characteristic for Malaria RDTs. We’ll use this to then find the
surveys that include this characteristic. We can also at this point
filter for our desired countries and years. The DHS API allows for
countries to be filtered using by their countryIds, which is
one of the arguments in dhs_surveys(). To have a look at
what each countries countryId is we can use another of the API endpoints
first:
## what are the countryIds
ids <- dhs_countries(returnFields=c("CountryName", "DHS_CountryCode"))
str(ids)## 'data.frame': 91 obs. of 2 variables:
## $ DHS_CountryCode: chr "AF" "AL" "AO" "AM" ...
## $ CountryName : chr "Afghanistan" "Albania" "Angola" "Armenia" ...
# lets find all the surveys that fit our search criteria
survs <- dhs_surveys(surveyCharacteristicIds = 89,countryIds = c("CD","TZ"),surveyYearStart = 2013)
# and lastly use this to find the datasets we will want to download and let's download the flat files (.dat) datasets (have a look in the dhs_datasets documentation for all argument options, and fileformat abbreviations etc.)
datasets <- dhs_datasets(surveyIds = survs$SurveyId, fileFormat = "flat")
str(datasets)## 'data.frame': 19 obs. of 13 variables:
## $ FileFormat : chr "Flat ASCII data (.dat)" "Flat ASCII data (.dat)" "Flat ASCII data (.dat)" "Flat ASCII data (.dat)" ...
## $ FileSize : int 198561 7030083 3226262 8028957 11426382 4794941 1569680 6595349 63022 996906 ...
## $ DatasetType : chr "HIV Datasets" "Survey Datasets" "Survey Datasets" "Survey Datasets" ...
## $ SurveyNum : int 421 421 421 421 421 421 421 421 421 421 ...
## $ SurveyId : chr "CD2013DHS" "CD2013DHS" "CD2013DHS" "CD2013DHS" ...
## $ FileType : chr "HIV Test Results Recode" "Births Recode" "Couples' Recode" "Household Recode" ...
## $ FileDateLastModified: chr "November, 14 2014 12:48:34" "November, 17 2014 15:42:54" "November, 17 2014 15:43:04" "September, 19 2016 09:57:20" ...
## $ SurveyYearLabel : chr "2013-14" "2013-14" "2013-14" "2013-14" ...
## $ SurveyType : chr "DHS" "DHS" "DHS" "DHS" ...
## $ SurveyYear : int 2013 2013 2013 2013 2013 2013 2013 2013 2013 2013 ...
## $ DHS_CountryCode : chr "CD" "CD" "CD" "CD" ...
## $ FileName : chr "CDAR61FL.ZIP" "CDBR61FL.ZIP" "CDCR61FL.ZIP" "CDHR61FL.ZIP" ...
## $ CountryName : chr "Congo Democratic Republic" "Congo Democratic Republic" "Congo Democratic Republic" "Congo Democratic Republic" ...Lastly, we recommended to download either the spss (.sav),
fileFormat = "SV", or the flat file (.dat),
fileFormat = "FL" datasets. The flat is quicker, but there
are still one or two datasets that don’t read correctly, whereas the
.sav files are slower to read in but so far no datasets have been found
that don’t read in correctly.
We can now use this to download our datasets for further analysis.
3. Download survey datasets
We can now go ahead and download our datasets. To do this we need to
first create a client. The client is an R6 class (similar
to R’s built in reference classes and make caching survey and API
queries more reproducible) and will be used to log in to your DHS
account, download datasets for you, and help query those datasets for
the question you are interested in. The client will also cache all of
these processes, which really helps increase the reproducibility of your
analysis.
In order to set up our credentials we use the function
set_rdhs_config(). This will require providing as arguments
your email and project for which you want to
download datasets from. You will then be prompted for your password.
You can also specify a directory for datasets and API calls to be
cached to using cache_path. If you do not provide an
argument for cache_path you will be prompted to provide
permission to rdhs to save your datasets and API calls
within your user cache directory for your operating system. This is to
comply with CRAN’s requests for permission to be granted before writing
to system files. If you do not grant permission, these will be written
within your R temporary directory (as we saw above when we first used
one of the functions to query the API). Similarly if you do not also
provide an argument for config_path, this will be saved
within your temp directory unless permission is granted. Your config
files will always be called “rdhs.json”, so that rdhs can
find them easily.
## create a client
## set up your credentials
config <- set_rdhs_config(email = "rdhs.tester@gmail.com",
project = "Testing Malaria Investigations")
client <- client_dhs(config)
client## <client_dhs>
## Public:
## available_datasets: function (clear_cache_first = FALSE)
## clear_namespace: function (namespace)
## dhs_api_request: function (api_endpoint, query = list(), api_key = private$api_key,
## extract: function (questions, add_geo = FALSE)
## get_cache_date: function ()
## get_config: function ()
## get_datasets: function (dataset_filenames, download_option = "rds", reformat = FALSE,
## get_downloaded_datasets: function ()
## get_root: function ()
## get_variable_labels: function (dataset_filenames = NULL, dataset_paths = NULL, rm_na = FALSE)
## initialize: function (config, api_key = NULL, root = rappdirs_rdhs())
## save_client: function ()
## set_cache_date: function (date)
## survey_questions: function (dataset_filenames, search_terms = NULL, essential_terms = NULL,
## survey_variables: function (dataset_filenames, variables, essential_variables = NULL,
## Private:
## api_endpoints: data indicators countries surveys surveycharacteristics ...
## api_key: ********
## cache_date: 2018-09-24 16:36:45
## check_available_datasets: function (filenames)
## config: rdhs_config
## na_s: ^na -|^na-|.*-na$|.* - na$| \{NA\}$|.* NA$|.*NA$
## package_version: package_version, numeric_version
## root: C:\Users\Oliver\AppData\Local\Oliver\rdhs\Cache
## storr: storr, R6
## url: https://api.dhsprogram.com/rest/dhs/
## user_declared_root: NULLBefore we use our client to download our datasets, it is worth
mentioning that the client can be passed as an argument to any of the
API functions we have just seen. This will then cache the results for
you, so that if you are working remotely or without a good internet
connection you can still return your previous API requests. This is what
is happening behind the scenes anyway, as rdhs will create
a client for you when you call an API function (if one does not already
exist).
Now back to our dataset downloads. If we have a look back at our datasets object, we’ll see there are 19 datasets listed. However, not all of them will be relevant to our malaria RDT questions. One approach is to head to the DHS website and have a look at the DHS Recodes, and look at the recodes that relate to the surveys. The other alternative is to download all the surveys and then query the variables within them. This is what we’ll demonstrate here as it also demonstrates more of the package’s functionality:
So first we will download all these datasets:
# download datasets
downloads <- client$get_datasets(datasets$FileName)The function returns a list with a file path to where the downloaded
datasets have been saved to. By default the files will download quietly,
i.e. no progress is shown. However, if you want to see the progress then
you can control this by setting this in your config using the
verbose_download argument.
4. Load datasets and associated metadata into R.
We can now examine what it is we have actually downloaded, by reading in one of these datasets:
# read in our dataset
cdpr <- readRDS(downloads$CDPR61FL)The dataset returned here contains all the survey questions within the dataset, and what their survey variables are:
# let's look at the variable_names
head(get_variable_labels(cdpr))## variable description
## 1 hhid Case Identification
## 2 hvidx Line number
## 3 hv000 Country code and phase
## 4 hv001 Cluster number
## 5 hv002 Household number
## 6 hv003 Respondent's line number (answering Household questionnaire)
# and then the dataset
class(cdpr$hv024)## [1] "labelled"This is the default behaviour for the client function
get_datasets - it will download the datasets for you, and
then by default save them in your client’s root directory and then unzip
them and read them in for you, and save the resultant data.frame as a
.rds object within the client’s root directory. You can control this
behaviour using the download_option argument as such:
-
client$get_datasets(download_option = "zip")- Just the downloaded zip will be saved -
client$get_datasets(download_option = "rds")- Just the read in rds will be saved -
client$get_datasets(download_option = "both")- The zip is downloaded and saved as well as the read in rds
The other main reason for reading the dataset in straight away as the default option is that the created table of all the survey variables and their definitions is cached then and there, which then allows us to quickly query for particular search terms or survey variables:
# rapid diagnostic test search
questions <- client$survey_questions(datasets$FileName, search_terms = "malaria rapid test")
table(questions$dataset_filename)##
## CDHR61FL CDPR61FL TZHR7AFL TZPR7AFL
## 24 1 48 1What we see from the questions is that the question “Result of malaria rapid test” appears in a few different datasets. This is because the household member recode datasets (CDPR61SV, TZPR7HSV) stores information about the children in a household, with one row per child, whereas the household recode (CDHR61SV, TZHR7HSV) stores information about the household, and thus flattens the information from each child into different subvariables (hml35$01/02 etc). As such it is easier to extract this information from the household member recodes.
5. Extract variables and combining datasets for pooled multi-survey analyses.
To extract our data we pass our questions object to the client
function extract, which will create a list with each
dataset and its extracted data as a data.frame. We also
have the option to add any geographic data available, which will
download the geographic data files for you and add this data to you
resultant extract:
# let's just use the PR files thus
datasets <- dhs_datasets(surveyIds = survs$SurveyId, fileFormat = "FL", fileType = "PR")
downloads <- client$get_datasets(datasets$FileName)
# and grab the questions from this again along with also questions detailing the province
questions <- client$survey_questions(datasets$FileName, search_terms = c("malaria rapid test"))
# and now extract the data
extract <- client$extract(questions, add_geo = FALSE)
# what does our extract look like
str(extract)## List of 2
## $ CDPR61FL:Classes 'dhs_dataset' and 'data.frame': 95949 obs. of 2 variables:
## ..$ hml35 : 'labelled' int [1:95949] NA NA NA NA NA NA NA 1 0 NA ...
## .. ..- attr(*, "label")= chr "Result of malaria rapid test"
## .. ..- attr(*, "labels")= Named int [1:3] 0 1 9
## .. .. ..- attr(*, "names")= chr [1:3] "negative" "positive" "missing"
## ..$ SurveyId: chr [1:95949] "CD2013DHS" "CD2013DHS" "CD2013DHS" "CD2013DHS" ...
## $ TZPR7AFL:Classes 'dhs_dataset' and 'data.frame': 64880 obs. of 2 variables:
## ..$ hml35 : 'labelled' int [1:64880] NA NA NA NA NA NA NA 0 NA NA ...
## .. ..- attr(*, "label")= chr "Result of malaria rapid test"
## .. ..- attr(*, "labels")= Named int [1:3] 0 1 9
## .. .. ..- attr(*, "names")= chr [1:3] "negative" "positive" "missing"
## ..$ SurveyId: chr [1:64880] "TZ2015DHS" "TZ2015DHS" "TZ2015DHS" "TZ2015DHS" ...The resultant extract is a list, with a new element for each
different dataset that you have extracted. The responses from the
dataset are by default stored as a labelled class from the haven package. This class
preserves the original semantics and can easily be coerced to factors
with haven::as_factor(). Special missing values are also
preserved. For more info on the labelled class have a look at
their github.
We can also query our datasets for the survey question variables. In
the example above the survey question was Result of malaria rapid
test and the variable was hml35. So if you knew the survey
variables that you wanted (either by looking at the Recode file or by
looking through the variable_names included in the datasets)
then we could search against these. So let’s grab the regions using
hv024 using the client function
survey_variables():
# and grab the questions from this now utilising the survey variables
questions <- client$survey_variables(datasets$FileName, variables = c("hv024","hml35"))
# and now extract the data
extract2 <- client$extract(questions, add_geo = FALSE)
# quick check
head(extract2$CDPR61FL)## hv024 hml35 SurveyId
## 1 4 NA CD2013DHS
## 2 4 NA CD2013DHS
## 3 4 NA CD2013DHS
## 4 4 NA CD2013DHS
## 5 4 NA CD2013DHS
## 6 4 NA CD2013DHS
head(extract2$TZPR7HFL)## NULL
# and just to prove that hml35 did actually read in okay (there are just lots of NA)
table(extract2$CDPR61FL$hml35,useNA = "always")##
## 0 1 9 <NA>
## 5260 2959 8 87722We can now combine our two dataframes for further analysis using the
rdhs package function rbind_labelled(). This
function works specifically with our lists of labelled data.frames:
# first let's bind our first extraction, without the hv024
extract_bound <- rbind_labelled(extract)
head(extract_bound)## hml35 SurveyId DATASET
## CDPR61FL.1 NA CD2013DHS CDPR61FL
## CDPR61FL.2 NA CD2013DHS CDPR61FL
## CDPR61FL.3 NA CD2013DHS CDPR61FL
## CDPR61FL.4 NA CD2013DHS CDPR61FL
## CDPR61FL.5 NA CD2013DHS CDPR61FL
## CDPR61FL.6 NA CD2013DHS CDPR61FL
# now let's try our second extraction
extract2_bound <- rbind_labelled(extract2)## Warning in rbind_labelled(extract2): Some variables have non-matching value labels: hv024.
## Inheriting labels from first data frame with labels.This hasn’t quite done what we might want in the second instance. The
hv024 variable stores the regions for these 2 countries, which
will not be the same and thus the labels will be different between the
two of them. Without specifying any additional arguments
rbind_labelled() will simply use the first data.frames
labelling as the default, which will mean that some of the Tanzanian
provinces will have been encoded as DRC provinces - not good! (This is a
similar problem in nature to say trying to add new character strings to
a factored data.frame).
There are a few work arounds. Firstly, we can specify a labels argument to the function which will detail how we should handle different variables. labels is a names list that specifies how to handle each variable. If we simply want to keep all the labels then we us the string “concatenate”:
# lets try concatenating the hv024
better_bound <- rbind_labelled(extract2, labels = list("hv024"="concatenate"))
head(better_bound$hv024)## <Labelled integer>
## [1] 6 6 6 6 6 6
##
## Labels:
## value label
## 1 arusha
## 2 bandundu
## 3 bas-congo
## 4 dar es salaam
## 5 dodoma
## 6 equateur
## 7 geita
## 8 iringa
## 9 kagera
## 10 kasai-occidental
## 11 kasai-oriental
## 12 kaskazini pemba
## 13 kaskazini unguja
## 14 katanga
## 15 katavi
## 16 kigoma
## 17 kilimanjaro
## 18 kinshasa
## 19 kusini pemba
## 20 kusini unguja
## 21 lindi
## 22 maniema
## 23 manyara
## 24 mara
## 25 mbeya
## 26 mjini magharibi
## 27 morogoro
## 28 mtwara
## 29 mwanza
## 30 njombe
## 31 nord-kivu
## 32 orientale
## 33 pwani
## 34 rukwa
## 35 ruvuma
## 36 shinyanga
## 37 simiyu
## 38 singida
## 39 sud-kivu
## 40 tabora
## 41 tangaWe could also specify new labels for a variable. For example, imagine
the two datasets encoded their RDT responses differently, with the first
one as c("No","Yes") and the other as
c("Negative","Positive"). These would be for our purposes
the same response, and so we could either leave it and all our results
would use the c("No","Yes") labelling. But we may want to
use the latter as it’s more informative/correct, or we may want to be
crystal clear and use c("NegativeTest","PositiveTest"). we
can do that like this:
# lets try concatenating the hv024 and providing new labels
better_bound <- rbind_labelled(extract2,
labels = list("hv024"="concatenate",
"hml35"=c("NegativeTest"=0, "PositiveTest"=1)))
# and our new label
head(better_bound$hml35)## <Labelled integer>
## [1] NA NA NA NA NA NA
##
## Labels:
## value label
## 0 NegativeTest
## 1 PositiveTestThe other option is to not use the labelled class at all. We can
control this when we download our datasets, using the argument
reformat=TRUE. This will ensure that no factors or labels
are used and it is just the raw data. When this option is set the object
returned by client$get_datasets() no longer has any
labelled classes or factors. However, we can still recover the variable
table for a dataset using get_variable_labels(), which will
take any dataset output by get_datasets() and return a
data.frame describing the survey question variables and definitions.
# download the datasets with the reformat arguments
downloads <- client$get_datasets(datasets$FileName, reformat=TRUE)
# grab the questions but specifying the reformat argument
questions <- client$survey_variables(datasets$FileName, variables = c("hv024", "hml35"),
reformat=TRUE)
# and now extract the data
extract3 <- client$extract(questions, add_geo = FALSE)
# group our results
bound_no_labels <- rbind_labelled(extract3)
# what does our hv024 look like now
class(bound_no_labels$hv024[1])## [1] "character"The hv024 column is now just characters, which is possibly the best option depending on your downstream analysis/preferences. It’s for this reason that the geographic data that is added is never turned into factors or labels.
Lastly, we can now use our extract dataset to carry out some regression analysis, to investigate the relationship between malaria prevalence and the quality of wall materials. To do this we will need to first grab the sample weights and stratification from the surveys, along with the extra variables and we will then check the RDT prevalence calculated using the raw data versus the API:
# grab the additional variable hv023 and hv024 which have the strata and weights respectively
questions <- client$survey_variables(datasets$FileName,variables = c("hv005","hv021","hv022","hv023","hv024",
"hv025","hv214","hml20", "hc1","hml35"))
extraction <- client$extract(questions,TRUE)
# now concatenate the provinces as before and remove missing responses
dat <- rbind_labelled(extraction,labels=list("hv024"="concatenate","hv214"="concatenate"))
dat <- dat[-which(dat$hml35==9),] # remove missing responses
# and we are going to compare our extract to the API malaria prevalence by RDT, which is for those between 6 and 59 months
dat <- dat[-which(!dat$hc1>6 & dat$hc1<=60),]
# create a denominator response for hml35
dat$hml35denom <- as.integer(!is.na(dat$hml35))
dat$bricks <- dat$hv214 %in% c(8,18,5,9,10)
dat$net <- as.logical(dat$hml20)
# specify the strata and sample weights
dat$strata <- paste0(dat$hv023,dat$DATASET)
dat$hv005 <- dat$hv005/1e6
# construct a survey design using the survey pacakge
library(survey)
# construct the sample design and calculate the mean and totals
des <- survey::svydesign(~CLUSTER+DATASET,data=dat,weight=~hv005)
results <- cbind(survey::svyby(~hml35,by=~DHSREGNA+DATASET, des, survey::svyciprop,na.rm=TRUE),
survey::svyby(~hml35denom,by=~DHSREGNA+DATASET, des, survey::svytotal,na.rm=TRUE))
results <- results[order(results$DATASET),]
# grab the same data from the API
dhs_api_data <- dhs_data(countryIds = c("CD","TZ"),indicatorIds = "ML_PMAL_C_RDT",breakdown = "subnational",surveyYearStart = 2013, surveyYearEnd = 2016)
dhs_api_data <- cbind(dhs_api_data$Value,dhs_api_data$DenominatorWeighted,dhs_api_data$CharacteristicLabel, dhs_api_data$SurveyId)
api <- dhs_api_data[!grepl("\\.\\.",dhs_api_data[,3]),] # remove subregions included in Tanzania
api <- api[order(apply(api[,4:3],1,paste,collapse="")),]
# bind the results and remove duplicate Region Columns
comparison <- cbind(results[,c(1,3,7)],api[])
names(comparison) <- c("Region","Survey_RDT_Prev","Survey_RDT_Denom","API_RDT_Prev","API_RDT_Denom","API_Regions","SurveyID")
head(comparison[,c(1,2,4,3,5,7)])## Region Survey_RDT_Prev API_RDT_Prev
## Bandundu.CDPR61FL Bandundu 0.2038607 20.2
## Bas-Congo.CDPR61FL Bas-Congo 0.4761599 47.1
## Equateur.CDPR61FL Equateur 0.2732268 27.4
## Kasai-Occidental.CDPR61FL Kasai-Occidental 0.4507341 44.5
## Kasai-Oriental.CDPR61FL Kasai-Oriental 0.4923113 49.4
## Katanga.CDPR61FL Katanga 0.3989890 38.9
## Survey_RDT_Denom API_RDT_Denom SurveyID
## Bandundu.CDPR61FL 1415.6056 1414 CD2013DHS
## Bas-Congo.CDPR61FL 342.6473 347 CD2013DHS
## Equateur.CDPR61FL 1267.4746 1236 CD2013DHS
## Kasai-Occidental.CDPR61FL 604.6050 612 CD2013DHS
## Kasai-Oriental.CDPR61FL 892.6006 894 CD2013DHS
## Katanga.CDPR61FL 861.2030 844 CD2013DHSIt’s a little off, which will be due to the specific stratification the DHS Program will have used, as well as potentially how they have grouped the primary sampling units. We are hoping to get this information from the DHS for each survey so we can make this process more streamline again.
And lastly we will construct a logistic regression to investigate the
relationship between a positive malaria RDT and whether the main walls
of an individual’s house were made of bricks or similar, while adjusting
for urban/rural (v025) and fixed effects for each
survey.
# contsruct our glm using svyglm and specify quasibinomial to handle the na in hml35
summary(svyglm(hml35 ~ DATASET + hv025 + net + bricks, des, family="quasibinomial"))##
## Call:
## svyglm(formula = hml35 ~ DATASET + hv025 + net + bricks, des,
## family = "quasibinomial")
##
## Survey design:
## survey::svydesign(~CLUSTER + DATASET, data = dat, weight = ~hv005)
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.65528 0.23267 -7.114 3.20e-12 ***
## DATASETTZPR7AFL -0.95553 0.12901 -7.406 4.39e-13 ***
## hv025 0.52558 0.13009 4.040 6.03e-05 ***
## netTRUE 0.06529 0.07163 0.911 0.362
## bricksTRUE -0.72109 0.13517 -5.335 1.36e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for quasibinomial family taken to be 0.9563804)
##
## Number of Fisher Scoring iterations: 4What we can see is that a significant negative gradient was associated with walls being made of bricks or similarly good materials in comparison to malaria positivity rates by RDT. What is also interesting is that whether the individual slept under a long lasting insecticidal net (hml20 that we converted to net) was not significant.
Summary and further thoughts
Hopefully the above tutorial has shown how the rdhs
package can facilitate both querying the DHS API and hopefully make
downloading and interacting with the raw datasets a smoother, more
reproducible process. It is worth bearing in mind though, that creating
a harmonised dataset is not always as easy as the example above - a lot
of the time survey variables differ across years and surveys, which is
hopefully when the survey_questions functionality will make
it easier to first filter down to those that include the relevant
questions before having to decide which survey questions are valid.
Any suggestions or comments/corrections/errors/ideas please let me know either in the issues or send me an email at “o.watson15@imperial.ac.uk”. And if there is any further functionality that you think you would be useful, then also let me know. :)