The goal of GLMMcosinor is to fit a cosinor model to rhythmic data except with all the flexibility and functionality of a generalised linear (mixed-) model (GLM) framework using glmmTMB.
For an introduction to the cosinor model, see the getting started vignette.
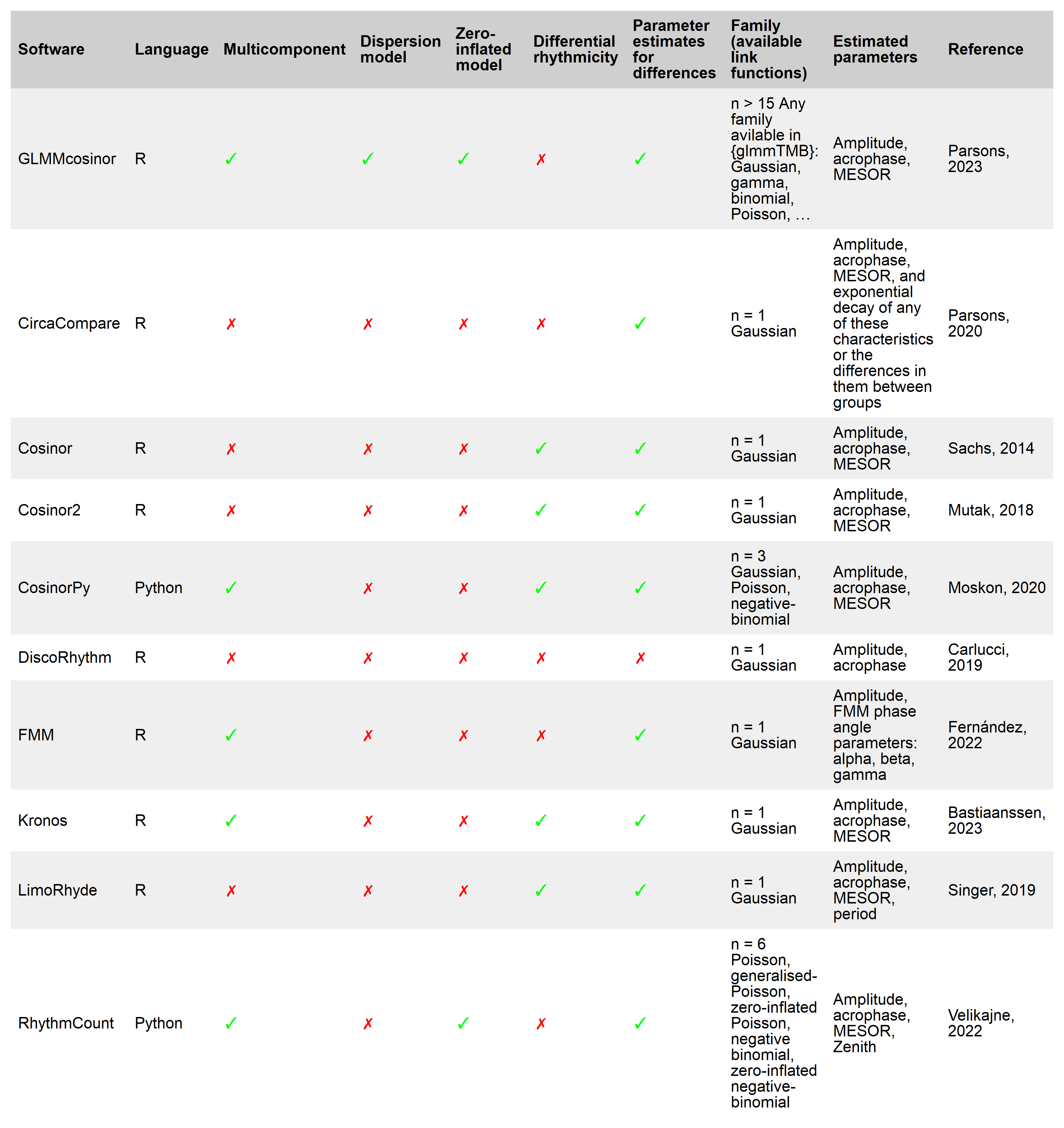
Existing statistical software for circadian data analyses (including cosinor or circacompare) allow the user to fit data using a regression model, but many are limited due to their inability to specify a link function, multiple components, or a hierarchical structure. GLMMcosinor aims to be comprehensive and flexible and is an improvement on other implementations of the cosinor model in R or Python. See table below for features available within currently available methods.
Installation
You can install the development version of GLMMcosinor from GitHub with:
# install.packages("remotes")
remotes::install_github("RWParsons/GLMMcosinor")Example
This is a basic example which shows you how to solve a common problem:
model <- cglmm(
vit_d ~ X + amp_acro(time, group = "X", period = 12),
data = vitamind
)
#> Warning in checkMatrixPackageVersion(): Package version inconsistency detected.
#> TMB was built with Matrix version 1.6.1.1
#> Current Matrix version is 1.5.4.1
#> Please re-install 'TMB' from source using install.packages('TMB', type = 'source') or ask CRAN for a binary version of 'TMB' matching CRAN's 'Matrix' package
summary(model)
#>
#> Conditional Model
#> Raw model coefficients:
#> estimate standard.error lower.CI upper.CI p.value
#> (Intercept) 29.6897959 0.4583696 28.7914079 30.58818 < 2.22e-16 ***
#> X1 1.9018623 0.7919688 0.3496320 3.45409 0.016331 *
#> X0:main_rrr1 0.9307876 0.6260656 -0.2962784 2.15785 0.137087
#> X1:main_rrr1 6.5102900 0.9303406 4.6868560 8.33372 2.6011e-12 ***
#> X0:main_sss1 6.2009896 0.6701952 4.8874311 7.51455 < 2.22e-16 ***
#> X1:main_sss1 4.8184618 0.8963299 3.0616875 6.57524 7.6257e-08 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Transformed coefficients:
#> estimate standard.error lower.CI upper.CI p.value
#> (Intercept) 29.6897959 0.4583696 28.7914079 30.58818 < 2.22e-16 ***
#> [X=1] 1.9018623 0.7919688 0.3496320 3.45409 0.016331 *
#> [X=0]:amp1 6.2704576 0.6696564 4.9579551 7.58296 < 2.22e-16 ***
#> [X=1]:amp1 8.0994722 1.1028357 5.9379540 10.26099 2.0696e-13 ***
#> [X=0]:acr1 1.4218056 0.1000725 1.2256670 1.61794 < 2.22e-16 ***
#> [X=1]:acr1 0.6371544 0.1360809 0.3704408 0.90387 2.8385e-06 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
autoplot(model, superimpose.data = TRUE)
polar_plot(model)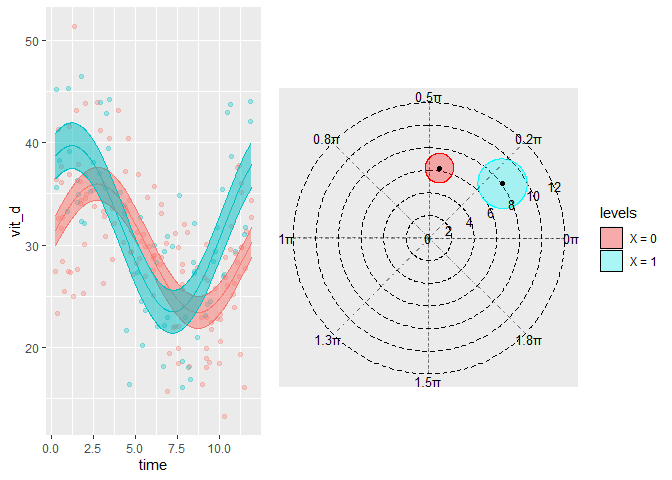
Citation
citation("GLMMcosinor")
#> Warning in citation("GLMMcosinor"): no date field in DESCRIPTION file of
#> package 'GLMMcosinor'
#>
#> To cite package 'GLMMcosinor' in publications use:
#>
#> Parsons R, Jayasinghe O, White N (2023). _GLMMcosinor: Fit A Cosinor
#> Model Using A Generalised Mixed Modelling Framework_.
#> https://github.com/RWParsons/GLMMcosinor,
#> https://rwparsons.github.io/GLMMcosinor/.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {GLMMcosinor: Fit A Cosinor Model Using A Generalised Mixed Modelling
#> Framework},
#> author = {Rex Parsons and Oliver Jayasinghe and Nicole White},
#> year = {2023},
#> note = {https://github.com/RWParsons/GLMMcosinor,
#> https://rwparsons.github.io/GLMMcosinor/},
#> }