rgbif tutorial
for v0.9.3
Seach and retrieve data from the Global Biodiverity Information Facilty (GBIF)
rgbif is an R package to search and retrieve data from the Global Biodiverity Information Facilty (GBIF). rgbif wraps R code around the GBIF API to allow you to talk to GBIF from R.
## Installation
Install `rgbif` from CRAN for more stable version
```r
install.packages("rgbif")
```
Or from Github for the development version
```r
devtools::install_github("ropensci/rgbif")
```
Load rgbif
```r
library("rgbif")
```
********************
## Usage
## Number of occurrences
Search by type of record, all observational in this case
```r
occ_count(basisOfRecord='OBSERVATION')
#> [1] 87417489
```
Records for **Puma concolor** with lat/long data (georeferened) only. Note that `hasCoordinate` in `occ_search()` is the same as `georeferenced` in `occ_count()`.
```r
occ_count(taxonKey=2435099, georeferenced=TRUE)
#> [1] 2878
```
All georeferenced records in GBIF
```r
occ_count(georeferenced=TRUE)
#> [1] 568769410
```
Records from Denmark
```r
denmark_code <- isocodes[grep("Denmark", isocodes$name), "code"]
occ_count(country=denmark_code)
#> [1] 10280120
```
Number of records in a particular dataset
```r
occ_count(datasetKey='9e7ea106-0bf8-4087-bb61-dfe4f29e0f17')
#> [1] 4591
```
All records from 2012
```r
occ_count(year=2012)
#> [1] 39022386
```
Records for a particular dataset, and only for preserved specimens
```r
occ_count(datasetKey='e707e6da-e143-445d-b41d-529c4a777e8b', basisOfRecord='OBSERVATION')
#> [1] 2120907
```
## Search for taxon names
Get possible values to be used in taxonomic rank arguments in functions
```r
taxrank()
#> [1] "kingdom" "phylum" "class" "order"
#> [5] "family" "genus" "species" "infraspecific"
```
`name_lookup()` does full text search of name usages covering the scientific and vernacular name, the species description, distribution and the entire classification across all name usages of all or some checklists. Results are ordered by relevance as this search usually returns a lot of results.
By default `name_lookup()` returns five slots of information: meta, data, facets, hierarchies, and names. hierarchies and names elements are named by their matching GBIF key in the `data.frame` in the data slot.
```r
out <- name_lookup(query='mammalia')
```
```r
names(out)
#> [1] "meta" "data" "facets" "hierarchies" "names"
```
```r
out$meta
#> offset limit endOfRecords count
#> 1 0 100 FALSE 147188
```
```r
head(out$data)
#> key scientificName
#> 1 115507497 Mammalia (awaiting allocation)
#> 2 359 Mammalia
#> 3 100375341 Mammalia
#> 4 113391223 Mammalia Linnaeus, 1758
#> 5 100348839 Mammalia Linnaeus 1758
#> 6 115507496 Mammalia (awaiting allocation)
#> datasetKey parentKey parent kingdom
#> 1 714c64e3-2dc1-4bb7-91e4-54be5af4da12 115507496 Mammalia Animalia
#> 2 d7dddbf4-2cf0-4f39-9b2a-bb099caae36c 44 Chordata Animalia
#> 3 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 NA
#> 4 cbb6498e-8927-405a-916b-576d00a6289b 115330302 Chordata Animalia
#> 5 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 100347572 Chordata
#> 6 714c64e3-2dc1-4bb7-91e4-54be5af4da12 115506762 Mammalia Animalia
#> phylum order family kingdomKey phylumKey classKey orderKey
#> 1 Chordata Mammalia Mammalia 115499364 115503274 115506762 115507496
#> 2 Chordata 1 44 359 NA
#> 3 NA NA 100375341 NA
#> 4 Chordata 112707351 115330302 113391223 NA
#> 5 Chordata NA 100347572 100348839 NA
#> 6 Chordata Mammalia 115499364 115503274 115506762 115507496
#> familyKey canonicalName authorship nameType taxonomicStatus rank
#> 1 115507497 Mammalia NO_NAME ACCEPTED FAMILY
#> 2 NA Mammalia SCIENTIFIC ACCEPTED CLASS
#> 3 NA Mammalia SCIENTIFIC CLASS
#> 4 NA Mammalia Linnaeus, 1758 SCIENTIFIC CLASS
#> 5 NA Mammalia Linnaeus, 1758 SCIENTIFIC CLASS
#> 6 NA Mammalia NO_NAME ACCEPTED ORDER
#> numDescendants numOccurrences habitats nomenclaturalStatus
#> 1 138 0 NA
#> 2 29630 0 MARINE NA
#> 3 0 0 NA
#> 4 3545 0 NA
#> 5 0 0 NA
#> 6 139 0 NA
#> threatStatuses synonym class nubKey extinct
#> 1 NA FALSE Mammalia NA NA
#> 2 NA FALSE Mammalia 359 FALSE
#> 3 NA FALSE Mammalia 359 TRUE
#> 4 NA FALSE Mammalia 359 NA
#> 5 NA FALSE Mammalia 359 NA
#> 6 NA FALSE Mammalia NA NA
```
```r
out$facets
#> NULL
```
```r
out$hierarchies[1:2]
#> $`115507497`
#> rankkey name
#> 1 115499364 Animalia
#> 2 115503274 Chordata
#> 3 115506762 Mammalia
#> 4 115507496 Mammalia
#>
#> $`359`
#> rankkey name
#> 1 1 Animalia
#> 2 44 Chordata
```
```r
out$names[2]
#> $`100375341`
#> vernacularName language
#> 1 Säugetiere deu
#> 2 Triconodont cat
#> 3 Triconodonta ces
#> 4 Triconodonta nld
#> 5 Triconodonta por
#> 6 Trykonodonty pol
#> 7 Триконодонты rus
```
Search for a genus
```r
head(name_lookup(query='Cnaemidophorus', rank="genus", return="data"))
#> key scientificName
#> 1 1858636 Cnaemidophorus Wallengren, 1862
#> 2 113100610 Cnaemidophorus Wallengren, 1862
#> 3 100555508 Cnaemidophorus Wallengren, 1862
#> 4 100465203 Cnaemidophorus Wallengren, 1862
#> 5 115196907 Cnaemidophorus
#> 6 115216121 Cnaemidophorus
#> datasetKey nubKey parentKey parent
#> 1 d7dddbf4-2cf0-4f39-9b2a-bb099caae36c 1858636 8863 Pterophoridae
#> 2 cbb6498e-8927-405a-916b-576d00a6289b NA 115216114 Pterophoridae
#> 3 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 1858636 100557623 Pterophoridae
#> 4 39653f3e-8d6b-4a94-a202-859359c164c5 1858636 100465147 Pterophoridae
#> 5 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 1858636 100557623 Pterophoridae
#> 6 cbb6498e-8927-405a-916b-576d00a6289b NA 115216114 Pterophoridae
#> kingdom phylum order family genus kingdomKey
#> 1 Animalia Arthropoda Lepidoptera Pterophoridae Cnaemidophorus 1
#> 2 Animalia Arthropoda Lepidoptera Pterophoridae Cnaemidophorus 112707351
#> 3 Lepidoptera Pterophoridae Cnaemidophorus NA
#> 4 Animalia Arthropoda Lepidoptera Pterophoridae Cnaemidophorus 100382406
#> 5 Lepidoptera Pterophoridae Cnaemidophorus NA
#> 6 Animalia Arthropoda Lepidoptera Pterophoridae Cnaemidophorus 112707351
#> phylumKey classKey orderKey familyKey genusKey canonicalName
#> 1 54 216 797 8863 1858636 Cnaemidophorus
#> 2 112710199 112780522 115213976 115216114 113100610 Cnaemidophorus
#> 3 NA NA 115196607 100557623 100555508 Cnaemidophorus
#> 4 100382501 100384963 100448306 100465147 100465203 Cnaemidophorus
#> 5 NA NA 115196607 100557623 115196907 Cnaemidophorus
#> 6 112710199 112780522 115213976 115216114 115216121 Cnaemidophorus
#> authorship nameType taxonomicStatus rank numDescendants
#> 1 Wallengren, 1862 SCIENTIFIC ACCEPTED GENUS 4
#> 2 Wallengren, 1862 SCIENTIFIC GENUS 2
#> 3 Wallengren, 1862 SCIENTIFIC GENUS 0
#> 4 Wallengren, 1862 SCIENTIFIC DOUBTFUL GENUS 1
#> 5 SCIENTIFIC ACCEPTED GENUS 1
#> 6 SCIENTIFIC ACCEPTED GENUS 1
#> numOccurrences extinct habitats nomenclaturalStatus threatStatuses
#> 1 0 FALSE NA NA
#> 2 0 NA NA NA
#> 3 0 NA NA NA
#> 4 0 NA NA NA
#> 5 0 NA NA NA
#> 6 0 NA NA NA
#> synonym class publishedIn accordingTo taxonID acceptedKey accepted
#> 1 FALSE Insecta NA
#> 2 FALSE Insecta NA
#> 3 FALSE NA
#> 4 FALSE Insecta NA
#> 5 FALSE NA
#> 6 FALSE Insecta NA
```
Search for the class mammalia
```r
head(name_lookup(query='mammalia', return = 'data'))
#> key scientificName
#> 1 115507497 Mammalia (awaiting allocation)
#> 2 359 Mammalia
#> 3 100375341 Mammalia
#> 4 113391223 Mammalia Linnaeus, 1758
#> 5 100348839 Mammalia Linnaeus 1758
#> 6 115507496 Mammalia (awaiting allocation)
#> datasetKey parentKey parent kingdom
#> 1 714c64e3-2dc1-4bb7-91e4-54be5af4da12 115507496 Mammalia Animalia
#> 2 d7dddbf4-2cf0-4f39-9b2a-bb099caae36c 44 Chordata Animalia
#> 3 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 NA
#> 4 cbb6498e-8927-405a-916b-576d00a6289b 115330302 Chordata Animalia
#> 5 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 100347572 Chordata
#> 6 714c64e3-2dc1-4bb7-91e4-54be5af4da12 115506762 Mammalia Animalia
#> phylum order family kingdomKey phylumKey classKey orderKey
#> 1 Chordata Mammalia Mammalia 115499364 115503274 115506762 115507496
#> 2 Chordata 1 44 359 NA
#> 3 NA NA 100375341 NA
#> 4 Chordata 112707351 115330302 113391223 NA
#> 5 Chordata NA 100347572 100348839 NA
#> 6 Chordata Mammalia 115499364 115503274 115506762 115507496
#> familyKey canonicalName authorship nameType taxonomicStatus rank
#> 1 115507497 Mammalia NO_NAME ACCEPTED FAMILY
#> 2 NA Mammalia SCIENTIFIC ACCEPTED CLASS
#> 3 NA Mammalia SCIENTIFIC CLASS
#> 4 NA Mammalia Linnaeus, 1758 SCIENTIFIC CLASS
#> 5 NA Mammalia Linnaeus, 1758 SCIENTIFIC CLASS
#> 6 NA Mammalia NO_NAME ACCEPTED ORDER
#> numDescendants numOccurrences habitats nomenclaturalStatus
#> 1 138 0 NA
#> 2 29630 0 MARINE NA
#> 3 0 0 NA
#> 4 3545 0 NA
#> 5 0 0 NA
#> 6 139 0 NA
#> threatStatuses synonym class nubKey extinct
#> 1 NA FALSE Mammalia NA NA
#> 2 NA FALSE Mammalia 359 FALSE
#> 3 NA FALSE Mammalia 359 TRUE
#> 4 NA FALSE Mammalia 359 NA
#> 5 NA FALSE Mammalia 359 NA
#> 6 NA FALSE Mammalia NA NA
```
Look up the species Helianthus annuus
```r
head(name_lookup(query = 'Helianthus annuus', rank="species", return = 'data'))
#> key scientificName datasetKey
#> 1 100336353 Helianthus annuus L. 16c3f9cb-4b19-4553-ac8e-ebb90003aa02
#> 2 113584542 Helianthus annuus L. cbb6498e-8927-405a-916b-576d00a6289b
#> 3 3119195 Helianthus annuus L. d7dddbf4-2cf0-4f39-9b2a-bb099caae36c
#> 4 103340289 Helianthus annuus fab88965-e69d-4491-a04d-e3198b626e52
#> 5 114910965 Helianthus annuus ee2aac07-de9a-47a2-b828-37430d537633
#> 6 115006874 Helianthus annuus L. b4af7484-5acd-4804-8211-d738f13832c7
#> nubKey parentKey parent kingdom order family
#> 1 3119195 115178966 Helianthus Plantae Asterales Asteraceae
#> 2 3119195 115390134 Helianthus Plantae Asterales Asteraceae
#> 3 3119195 3119134 Helianthus Plantae Asterales Asteraceae
#> 4 NA 103340270 Helianthus Viridiplantae Asterales Asteraceae
#> 5 3119195 114995002 Helianthus Plantae Asterales Asteraceae
#> 6 3119195 115006870 Helianthus Compositae
#> genus species kingdomKey classKey orderKey familyKey
#> 1 Helianthus Helianthus annuus 115177477 100328106 100336278 115178941
#> 2 Helianthus Helianthus annuus 113551056 NA 115388548 115388694
#> 3 Helianthus Helianthus annuus 6 220 414 3065
#> 4 Helianthus Helianthus annuus 102974832 NA 103311652 103311763
#> 5 Helianthus Helianthus annuus 114991342 114994101 114994711 114994731
#> 6 Helianthus NA NA NA 115005819
#> genusKey speciesKey canonicalName authorship nameType rank
#> 1 115178966 100336353 Helianthus annuus L. SCIENTIFIC SPECIES
#> 2 115390134 113584542 Helianthus annuus L. SCIENTIFIC SPECIES
#> 3 3119134 3119195 Helianthus annuus L. SCIENTIFIC SPECIES
#> 4 103340270 103340289 Helianthus annuus SCIENTIFIC SPECIES
#> 5 114995002 114910965 Helianthus annuus SCIENTIFIC SPECIES
#> 6 115006870 NA Helianthus annuus L. SCIENTIFIC SPECIES
#> numDescendants numOccurrences habitats nomenclaturalStatus
#> 1 0 0
#> 2 0 0
#> 3 26 0 TERRESTRIAL
#> 4 2 0
#> 5 0 0
#> 6 8 0
#> threatStatuses synonym class basionymKey
#> 1 NA FALSE Magnoliopsida NA
#> 2 NA FALSE NA
#> 3 NA FALSE Magnoliopsida 7566760
#> 4 NA FALSE NA
#> 5 NA FALSE Magnoliopsida NA
#> 6 NA FALSE NA
#> basionym phylum phylumKey taxonomicStatus
#> 1 NA
#> 2 NA
#> 3 Helianthus lenticularis Douglas Tracheophyta 7707728 DOUBTFUL
#> 4 Streptophyta 102986054
#> 5 Tracheophyta 114993928
#> 6 NA
#> extinct publishedIn accordingTo acceptedKey accepted
#> 1 NA NA
#> 2 NA NA
#> 3 FALSE NA
#> 4 NA NA
#> 5 NA NA
#> 6 NA NA
```
The function `name_usage()` works with lots of different name endpoints in GBIF, listed at [http://www.gbif.org/developer/species#nameUsages](http://www.gbif.org/developer/species#nameUsages).
```r
library("plyr")
out <- name_usage(key=3119195, language="FRENCH", data='vernacularNames')
head(out$data)
#> vernacularName language country sourceTaxonKey
#> 1 Gewöhnliche Sonnenblume deu DE 116782143
#> 2 Sonnenblume deu 101321447
#> 3 alizeti swa 101321447
#> 4 annual sunflower eng 102234356
#> 5 common sunflower eng 102234356
#> 6 girasol spa 101321447
#> source preferred
#> 1 NA
#> 2 GRIN Taxonomy NA
#> 3 GRIN Taxonomy NA
#> 4 Integrated Taxonomic Information System (ITIS) NA
#> 5 Integrated Taxonomic Information System (ITIS) NA
#> 6 GRIN Taxonomy NA
```
The function `name_backbone()` is used to search against the GBIF backbone taxonomy
```r
name_backbone(name='Helianthus', rank='genus', kingdom='plants')
#> $usageKey
#> [1] 3119134
#>
#> $scientificName
#> [1] "Helianthus L."
#>
#> $canonicalName
#> [1] "Helianthus"
#>
#> $rank
#> [1] "GENUS"
#>
#> $status
#> [1] "ACCEPTED"
#>
#> $confidence
#> [1] 97
#>
#> $matchType
#> [1] "EXACT"
#>
#> $kingdom
#> [1] "Plantae"
#>
#> $phylum
#> [1] "Tracheophyta"
#>
#> $order
#> [1] "Asterales"
#>
#> $family
#> [1] "Asteraceae"
#>
#> $genus
#> [1] "Helianthus"
#>
#> $kingdomKey
#> [1] 6
#>
#> $phylumKey
#> [1] 7707728
#>
#> $classKey
#> [1] 220
#>
#> $orderKey
#> [1] 414
#>
#> $familyKey
#> [1] 3065
#>
#> $genusKey
#> [1] 3119134
#>
#> $synonym
#> [1] FALSE
#>
#> $class
#> [1] "Magnoliopsida"
```
The function `name_suggest()` is optimized for speed, and gives back suggested names based on query parameters.
```r
head( name_suggest(q='Puma concolor') )
#> key canonicalName rank
#> 1 6164620 Puma concolor cougar SUBSPECIES
#> 2 6164600 Puma concolor coryi SUBSPECIES
#> 3 6164604 Puma concolor stanleyana SUBSPECIES
#> 4 6164610 Puma concolor hippolestes SUBSPECIES
#> 5 6164622 Puma concolor puma SUBSPECIES
#> 6 7193927 Puma concolor concolor SUBSPECIES
```
## Single occurrence records
Get data for a single occurrence. Note that data is returned as a list, with slots for metadata and data, or as a hierarchy, or just data.
Just data
```r
occ_get(key=766766824, return='data')
#> name key decimalLatitude decimalLongitude
#> 1 Coloeus monedula 766766824 59.4568 17.9054
#> issues
#> 1 depunl,gass84
```
Just taxonomic hierarchy
```r
occ_get(key=766766824, return='hier')
#> name key rank
#> 1 Animalia 1 kingdom
#> 2 Chordata 44 phylum
#> 3 Aves 212 class
#> 4 Passeriformes 729 order
#> 5 Corvidae 5235 family
#> 6 Coloeus 4852454 genus
#> 7 Coloeus monedula 6100954 species
```
All data, or leave return parameter blank
```r
occ_get(key=766766824, return='all')
#> $hierarchy
#> name key rank
#> 1 Animalia 1 kingdom
#> 2 Chordata 44 phylum
#> 3 Aves 212 class
#> 4 Passeriformes 729 order
#> 5 Corvidae 5235 family
#> 6 Coloeus 4852454 genus
#> 7 Coloeus monedula 6100954 species
#>
#> $media
#> list()
#>
#> $data
#> name key decimalLatitude decimalLongitude
#> 1 Coloeus monedula 766766824 59.4568 17.9054
#> issues
#> 1 depunl,gass84
```
Get many occurrences. `occ_get` is vectorized
```r
occ_get(key=c(766766824, 101010, 240713150, 855998194), return='data')
#> name key decimalLatitude decimalLongitude
#> 1 Coloeus monedula 766766824 59.4568 17.9054
#> 2 Platydoras armatulus 101010 NA NA
#> 3 Pelosina 240713150 -77.5667 163.5830
#> 4 Sciurus vulgaris 855998194 58.4068 12.0438
#> issues
#> 1 depunl,gass84
#> 2
#> 3 cdround,gass84
#> 4 depunl,gass84
```
## Search for occurrences
By default `occ_search()` returns a `dplyr` like output summary in which the data printed expands based on how much data is returned, and the size of your window. You can search by scientific name:
```r
occ_search(scientificName = "Ursus americanus", limit = 20)
#> Records found [7802]
#> Records returned [20]
#> No. unique hierarchies [1]
#> No. media records [18]
#> Args [scientificName=Ursus americanus, limit=20, offset=0, fields=all]
#> First 10 rows of data
#>
#> name key decimalLatitude decimalLongitude
#> 1 Ursus americanus 1229610216 44.06086 -71.92712
#> 2 Ursus americanus 1253300445 44.65481 -72.67270
#> 3 Ursus americanus 1249277297 35.76789 -75.80894
#> 4 Ursus americanus 1229610234 44.06062 -71.92692
#> 5 Ursus americanus 1253314877 49.25782 -122.82786
#> 6 Ursus americanus 1249284297 43.68723 -72.32891
#> 7 Ursus americanus 1249296297 39.08590 -105.24586
#> 8 Ursus americanus 1253317181 43.64214 -72.52494
#> 9 Ursus americanus 1257415362 44.32746 -72.41007
#> 10 Ursus americanus 1262389246 43.80871 -72.20964
#> .. ... ... ... ...
#> Variables not shown: issues (chr), datasetKey (chr), publishingOrgKey
#> (chr), publishingCountry (chr), protocol (chr), lastCrawled (chr),
#> lastParsed (chr), extensions (chr), basisOfRecord (chr), taxonKey
#> (int), kingdomKey (int), phylumKey (int), classKey (int), orderKey
#> (int), familyKey (int), genusKey (int), speciesKey (int),
#> scientificName (chr), kingdom (chr), phylum (chr), order (chr),
#> family (chr), genus (chr), species (chr), genericName (chr),
#> specificEpithet (chr), taxonRank (chr), dateIdentified (chr), year
#> (int), month (int), day (int), eventDate (chr), modified (chr),
#> lastInterpreted (chr), references (chr), identifiers (chr), facts
#> (chr), relations (chr), geodeticDatum (chr), class (chr), countryCode
#> (chr), country (chr), rightsHolder (chr), identifier (chr),
#> verbatimEventDate (chr), datasetName (chr), verbatimLocality (chr),
#> collectionCode (chr), gbifID (chr), occurrenceID (chr), taxonID
#> (chr), license (chr), catalogNumber (chr), recordedBy (chr),
#> http...unknown.org.occurrenceDetails (chr), institutionCode (chr),
#> rights (chr), eventTime (chr), identificationID (chr),
#> occurrenceRemarks (chr), infraspecificEpithet (chr),
#> coordinateUncertaintyInMeters (dbl), informationWithheld (chr)
```
Or to be more precise, you can search for names first, make sure you have the right name, then pass the GBIF key to the `occ_search()` function:
```r
key <- name_suggest(q='Helianthus annuus', rank='species')$key[1]
occ_search(taxonKey=key, limit=20)
#> Records found [30216]
#> Records returned [20]
#> No. unique hierarchies [1]
#> No. media records [12]
#> Args [taxonKey=3119195, limit=20, offset=0, fields=all]
#> First 10 rows of data
#>
#> name key decimalLatitude decimalLongitude
#> 1 Helianthus annuus 1249279611 34.04810 -117.79884
#> 2 Helianthus annuus 1248872560 37.81227 -8.82959
#> 3 Helianthus annuus 1248887127 38.53339 -8.94263
#> 4 Helianthus annuus 1249286909 32.58747 -97.10081
#> 5 Helianthus annuus 1253308332 29.67463 -95.44804
#> 6 Helianthus annuus 1248873088 38.53339 -8.94263
#> 7 Helianthus annuus 1262375813 29.82586 -95.45604
#> 8 Helianthus annuus 1262379231 34.04911 -117.80066
#> 9 Helianthus annuus 1265544678 32.58747 -97.10081
#> 10 Helianthus annuus 1262385911 32.78328 -96.70352
#> .. ... ... ... ...
#> Variables not shown: issues (chr), datasetKey (chr), publishingOrgKey
#> (chr), publishingCountry (chr), protocol (chr), lastCrawled (chr),
#> lastParsed (chr), extensions (chr), basisOfRecord (chr), taxonKey
#> (int), kingdomKey (int), phylumKey (int), classKey (int), orderKey
#> (int), familyKey (int), genusKey (int), speciesKey (int),
#> scientificName (chr), kingdom (chr), phylum (chr), order (chr),
#> family (chr), genus (chr), species (chr), genericName (chr),
#> specificEpithet (chr), taxonRank (chr), dateIdentified (chr), year
#> (int), month (int), day (int), eventDate (chr), modified (chr),
#> lastInterpreted (chr), references (chr), identifiers (chr), facts
#> (chr), relations (chr), geodeticDatum (chr), class (chr), countryCode
#> (chr), country (chr), rightsHolder (chr), identifier (chr),
#> verbatimEventDate (chr), datasetName (chr), verbatimLocality (chr),
#> collectionCode (chr), gbifID (chr), occurrenceID (chr), taxonID
#> (chr), license (chr), catalogNumber (chr), recordedBy (chr),
#> http...unknown.org.occurrenceDetails (chr), institutionCode (chr),
#> rights (chr), eventTime (chr), identificationID (chr),
#> infraspecificEpithet (chr), nomenclaturalCode (chr), institutionID
#> (chr), dataGeneralizations (chr), footprintWKT (chr), municipality
#> (chr), county (chr), language (chr), occurrenceStatus (chr),
#> footprintSRS (chr), ownerInstitutionCode (chr), identifiedBy (chr),
#> reproductiveCondition (chr), higherClassification (chr), collectionID
#> (chr), coordinateUncertaintyInMeters (dbl), occurrenceRemarks (chr),
#> informationWithheld (chr), elevation (dbl), elevationAccuracy (dbl),
#> stateProvince (chr), recordNumber (chr), locality (chr), type (chr)
```
Like many functions in `rgbif`, you can choose what to return with the `return` parameter, here, just returning the metadata:
```r
occ_search(taxonKey=key, return='meta')
#> offset limit endOfRecords count
#> 1 300 200 FALSE 30216
```
You can choose what fields to return. This isn't passed on to the API query to GBIF as they don't allow that, but we filter out the columns before we give the data back to you.
```r
occ_search(scientificName = "Ursus americanus", fields=c('name','basisOfRecord','protocol'), limit = 20)
#> Records found [7802]
#> Records returned [20]
#> No. unique hierarchies [1]
#> No. media records [18]
#> Args [scientificName=Ursus americanus, limit=20, offset=0,
#> fields=name,basisOfRecord,protocol]
#> First 10 rows of data
#>
#> name protocol basisOfRecord
#> 1 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 2 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 3 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 4 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 5 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 6 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 7 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 8 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 9 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 10 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> .. ... ... ...
```
Most parameters are vectorized, so you can pass in more than one value:
```r
splist <- c('Cyanocitta stelleri', 'Junco hyemalis', 'Aix sponsa')
keys <- sapply(splist, function(x) name_suggest(x)$key[1], USE.NAMES=FALSE)
occ_search(taxonKey=keys, limit=5)
#> Occ. found [7192170 (1174), 6173536 (57), 2498387 (775349)]
#> Occ. returned [7192170 (5), 6173536 (5), 2498387 (5)]
#> No. unique hierarchies [7192170 (1), 6173536 (1), 2498387 (1)]
#> No. media records [7192170 (5), 6173536 (1), 2498387 (5)]
#> Args [taxonKey=7192170,6173536,2498387, limit=5, offset=0, fields=all]
#> First 10 rows of data from 7192170
#>
#> name key decimalLatitude decimalLongitude issues
#> 1 Cyanocitta stelleri 1147228297 34.73360 -119.9871
#> 2 Cyanocitta stelleri 1147052100 39.61584 -120.5881 cdround
#> 3 Cyanocitta stelleri 1147243804 39.61584 -120.5881 cdround
#> 4 Cyanocitta stelleri 1147083657 39.61584 -120.5881 cdround
#> 5 Cyanocitta stelleri 1147179622 39.63799 -120.6085 cdround
#> Variables not shown: datasetKey (chr), publishingOrgKey (chr),
#> publishingCountry (chr), protocol (chr), lastCrawled (chr),
#> lastParsed (chr), extensions (chr), basisOfRecord (chr),
#> establishmentMeans (chr), taxonKey (int), kingdomKey (int), phylumKey
#> (int), classKey (int), orderKey (int), familyKey (int), genusKey
#> (int), speciesKey (int), scientificName (chr), kingdom (chr), phylum
#> (chr), order (chr), family (chr), genus (chr), species (chr),
#> genericName (chr), specificEpithet (chr), infraspecificEpithet (chr),
#> taxonRank (chr), continent (chr), stateProvince (chr), year (int),
#> month (int), day (int), eventDate (chr), modified (chr),
#> lastInterpreted (chr), references (chr), identifiers (chr), facts
#> (chr), relations (chr), geodeticDatum (chr), class (chr), countryCode
#> (chr), country (chr), institutionID (chr), county (chr), language
#> (chr), gbifID (chr), type (chr), catalogNumber (chr),
#> occurrenceStatus (chr), vernacularName (chr), institutionCode (chr),
#> rights (chr), behavior (chr), identifier (chr), verbatimEventDate
#> (chr), higherGeography (chr), nomenclaturalCode (chr), endDayOfYear
#> (chr), georeferenceVerificationStatus (chr), locality (chr),
#> verbatimLocality (chr), collectionCode (chr), occurrenceID (chr),
#> recordedBy (chr), startDayOfYear (chr), occurrenceRemarks (chr),
#> accessRights (chr)
```
********************
## Maps
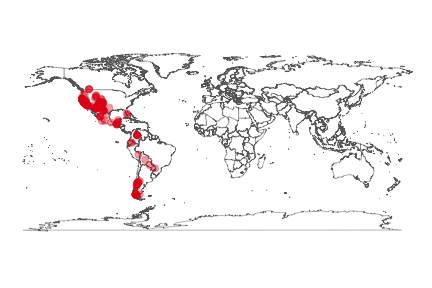
Static map using the ggplot2 package. Make a map of *Puma concolor* occurrences.
```r
key <- name_backbone(name='Puma concolor')$speciesKey
dat <- occ_search(taxonKey=key, return='data', limit=300)
gbifmap(dat)
```
## Citing
To cite `rgbif` in publications use:
> Scott Chamberlain, Carl Boettiger, Karthik Ram, Vijay Barve and Dan Mcglinn (2016). rgbif: Interface to the Global Biodiversity Information Facility API. R package version 0.9.3 https://github.com/ropensci/rgbif
## License and bugs
* License: [MIT](http://opensource.org/licenses/MIT)
* Report bugs at [our Github repo for rgbif](https://github.com/ropensci/rgbif/issues?state=open)
[Back to top](#top)
[gbifapi]: http://www.gbif.org/developer/summary
